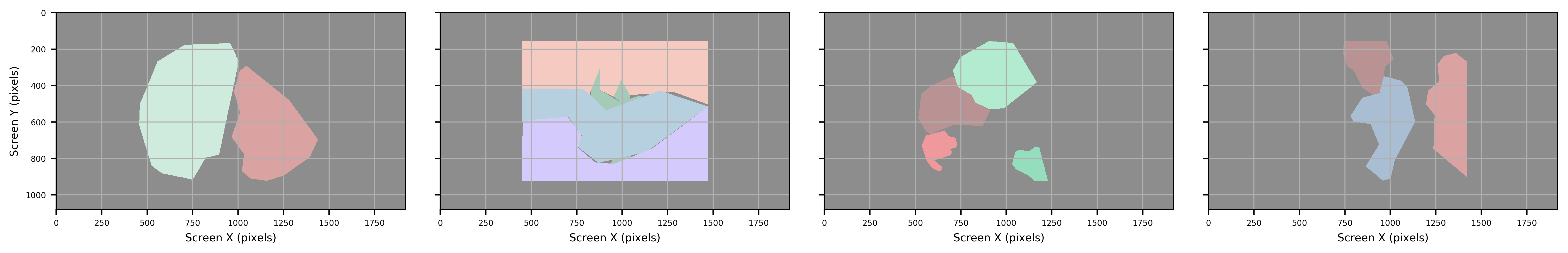
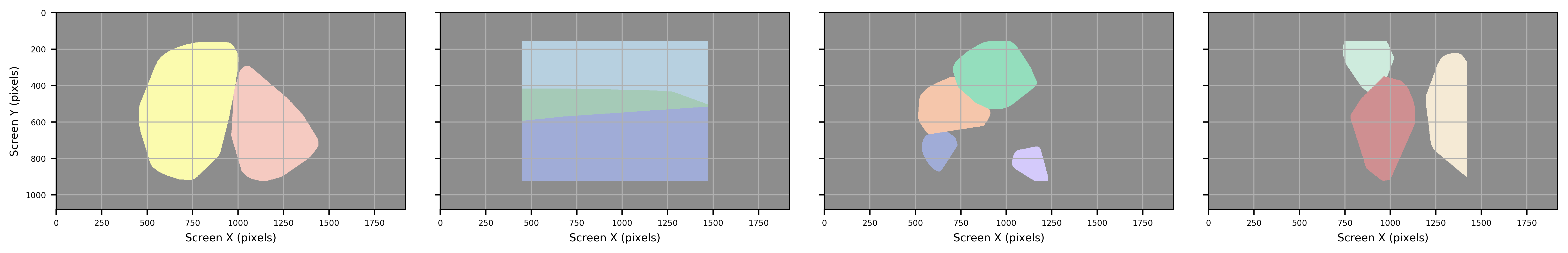
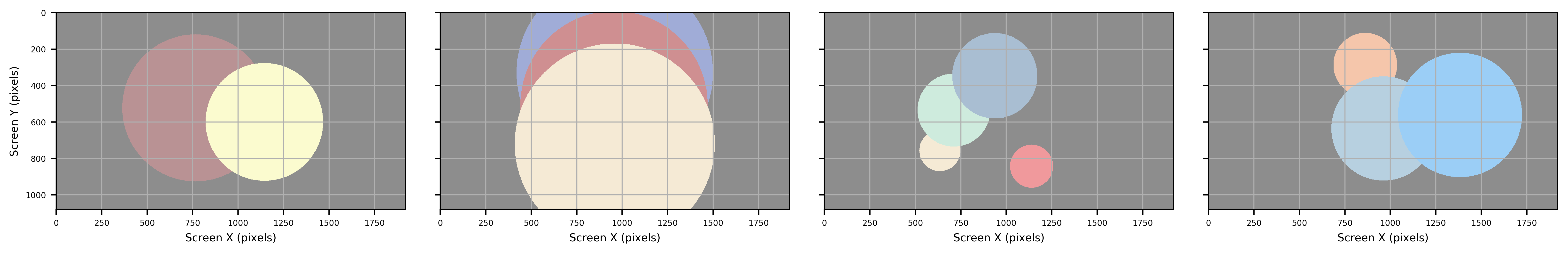
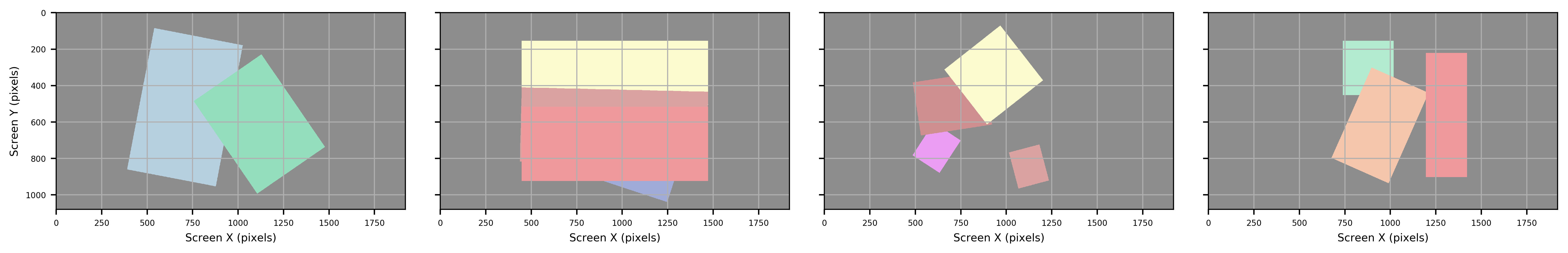
Region of Interest¶
imhr.eyetracking.ROI() can identify Region/Area of Interest using machine learning and image coding techniques.
Introduction¶
Example¶
[5]:
# import
import imhr
# parameters
path = imhr.__path__[0]
image_path = '%s/dist/roi/raw/2/'%(path)
output_path = '%s/dist/roi/output/'%(path)
metadata_source = '%s/dist/roi/raw/2/metadata.xlsx'%(path)
position = 'topcenter'
# initiate
roi = imhr.eyetracking.ROI(isMultiprocessing=False, isDebug=True,
# roi detection method and contour shape
detection='manual', shape='straight',
# path to read and export data
image_path=image_path, output_path=output_path, metadata_source=metadata_source,
# stimuli size and screen size
screensize=[1920,1080], recenter=[(1920*.50),(1080*.50)], scale=.75,
# applying new id, renaming variables and output
newcolumn={'position': position}, uuid=['image','roi','position'], append_output_name=position,
# image backend
image_backend='PIL')
# run
df, error = roi.process()
not multiprocessing
starting()
finished()
Parameters¶
Required Parameters¶
Here are all of the required parameters that are part of this module:
| Property | Description |
|---|---|
isMultiprocessing : bool |
Should the rois be generated using multiprocessing. Default is False. |
detection : str {‘manual’, ‘haarcascade’} |
How should the regions of interest be detected. Either manually (manual), through the use of highlighting layers in photo-editing software, or automatically through feature detection using haarcascade classifers from opencv. Default manual. |
image_path : str |
Image directory path. |
output_path : str |
Path to save data. |
roi_format : str {‘raw’, ‘dataviewer’, ‘both’} |
Format to export ROIs. Either to ‘csv’ (raw) or to Eyelink DataViewer ‘ias’ (dataviewer) or both (both). Default is both. Note: If roi_format = dataviewer, shape must be either be circle, rotated, or straight. |
metadata_source : str or None {‘path’, ‘embedded’} |
Metadata source. If metadata is being read from a spreadsheet, metadata_source should be equal to path the to the metadata file, else if metadata is embed within the image as a layer name, metadata_source = embedded. Although Photoshop PSD don’t directly provide support for metadata. However if each region of interest is stored as a seperate layer within a PSD, the layer name can be used to store metadata. To do this, the layer name has to be written as delimited text. Our code can read this data and extract relevant metadata. The delimiter can be either ; , | \t or \s (Delimiter type must be identified when running this code using the delimiter parameter. The default is ;.). Here’s an example using ; as a delimiter: >>> imagename = "BM001"; roiname = 1; feature = "lefteye"
Note: whitespace should be avoided from from each layer name. Whitespaces may cause errors during parsing. |
shape : str {‘polygon’, ‘hull’, ‘circle’, ‘rotated’, ‘straight’} |
Shape of machine readable boundaries for region of interest. Default is straight. polygon creates a Contour Approximation and will most closely match the orginal shape of the roi. hull creates a Convex Hull, which is similar to but not as complex as a Contour Approximation and will include bulges for areas that are convex. circle creates a mininum enclosing circle. Finally, both rotated and straight create a Bounding Rectangle, with the only difference being compensation for the mininum enclosing area for the box when using rotated. |
roicolumn : str |
The name of the label for the region of interest in your metadata. For example you may want to extract the column ‘feature’ from your metadata and use this as the label. Default is roi. |
uuid : list or None |
Create a unique id by combining a list of existing variables in the metadata. This is recommended if roi_format == dataviewer because of the limited variables allowed for ias files. Default is None. |
filetype: str {‘psd’, ‘tiff’, ‘dcm’, ‘png’, ‘bmp’, ‘jpg’} |
The filetype extension of the image file. Case insensitive. Default is psd. If psd, tiff or DICOM the file can be read as multilayered |
Optional Parameters¶
Here are optional parameters, not required to run the code, but allow output tweaking:
Additional core parameters
These properties control additional core parameters for the API:
| Property | Description |
|---|---|
cores : bool |
(if isMultiprocessing == True) Number of cores to use. Default is total available cores - 1. |
isLibrary : bool |
Check if required packages have been installed. Default is False. |
isDebug : bool |
Allow flags to be visible. Default is False. |
isDemo : bool |
Tests code with in-house images and metadata. Default is False. |
save_data : bool |
Save coordinates. Default is True. |
newcolumn : dict {str, str} or False |
Add additional column to metadata. This must be in the form of a dict in this form {key: value}. Default is False. |
save_raw_image : bool |
Save images. Default is True. |
append_output_name : bool or str |
Add appending name to all exported files (i.e. <’top_center’> IMG001_top_center.ias). Default is False. |
save_contour_image : bool |
Save generated contours as images. Default is True. |
scale : int |
If image is scaled during presentation, set scale. Default is 1. |
offset : list [int] |
Center point of image, relative to screensize. Default is [960, 540]. |
screensize : list [int] |
Monitor size is being presented. Default is [1920, 1080]. |
Data processing parameters
These properties control data is processed which include the type of haarcascade used, delimiters for metadata:
| Property | Description |
|---|---|
delimiter : str {‘;’ , ‘,’ , ‘|’ , ‘tab’ , ‘space’} |
(if source == psd) How is metadata delimited. Default is ;. |
classifiers : default or :obj:list of :obj:dict |
(if detection == haarcascade) Trained classifiers to use. Default is {‘eye_tree_eyeglasses’, ‘eye’, ‘frontalface_alt_tree’, ‘frontalface_alt’, ‘frontalface_alt2’,’frontalface_default’, ‘fullbody’, ‘lowerbody’, ‘profileface’, ‘smile’, ‘upperbody’}. Parameters are stored here. If you want to use custom classifiers, you can pass a list of classifiers and their arguments using the following format: >>> [{'custom_cascade': {
... 'file': 'haarcascade_eye.xml',
... 'type': 'profileface',
... 'path': './haarcascade_eye.xml',
... 'minN': 5,
... 'minS':(100,100),
... 'sF': 1.01 }
... }]
You can also pass custom arguments by calling them after initiation: >>> roi = imhr.eyetracking.ROI(detection='manual.....)
>>> roi.default_classifiers['eye']['minNeighbors'] = 10
|
Image export parameters
Here are properties specific to how images are exported after processing. The code can either use matplotlib
or PIL as a backend engine:
| Property | Description |
|---|---|
image_backend : str {‘matplotlib’, ‘PIL’} |
Backend for exporting image. Either matplotlib or PIL. Default is matplotlib. |
RcParams : bool |
A dictionary object including validation validating functions are defined and associated with rc parameters in class:matplotlib.RcParams. Default is None. |
background_color : list |
Set background color (RGB) for exporting images. Default is [110, 110, 110]. |
dpi : int or None |
(if save_image == True) Quality of exported images, refers to ‘dots per inch’. Default is 300. |
remove_axis : bool |
Remove axis from matplotlib.pyplot. Default is False. |
tight_layout : bool |
Remove whitespace from matplotlib.pyplot. Default is False. |
set_size_inches : bool |
Set size of matplotlib.pyplot according to screensize of ROI. Default is False. |
Creating ROI¶
There are two methods of creating regions of interest to be processed by the api: using manually coded images or haar cascades.
Manual coding¶
Using multilayered images (psd, dcm)
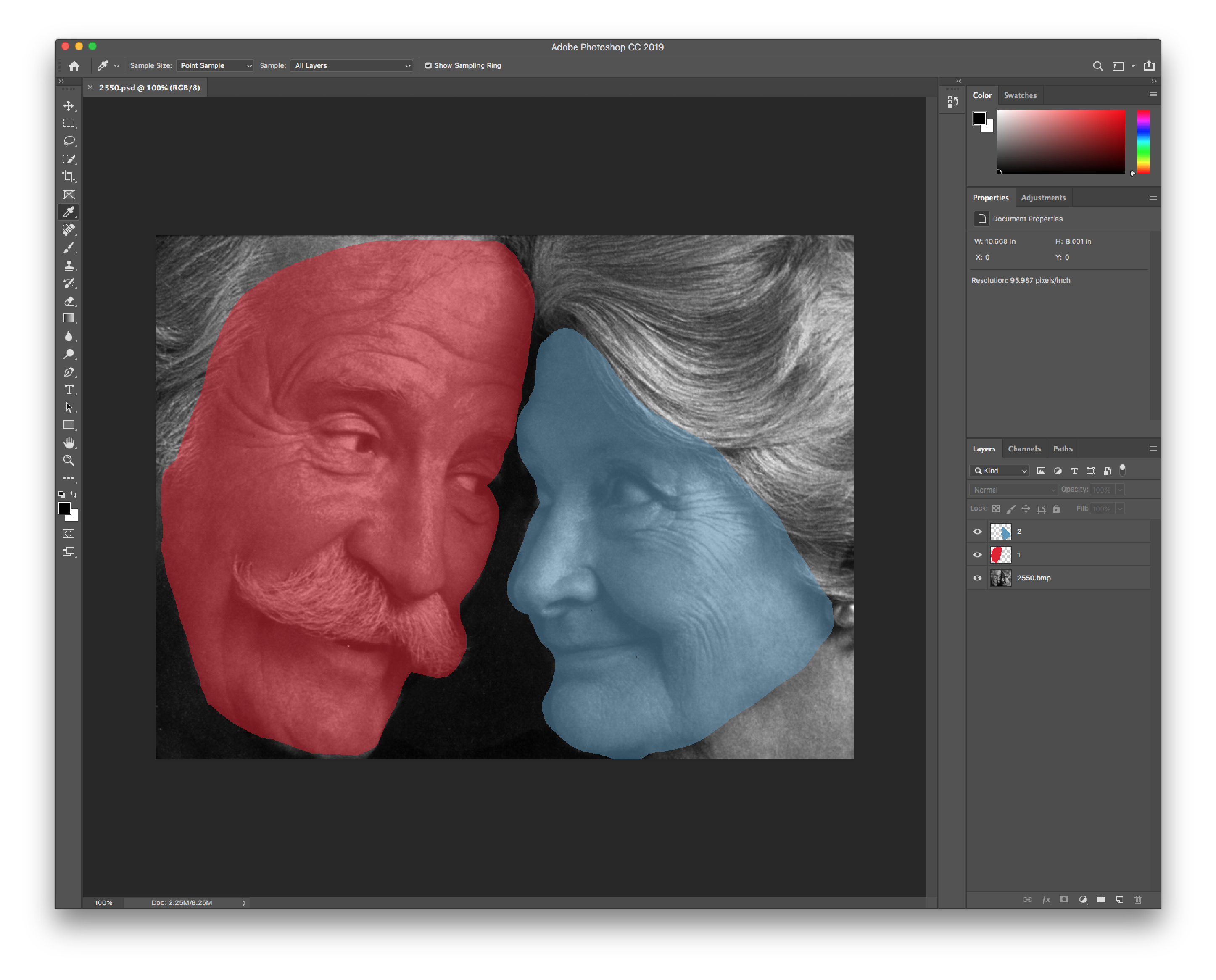
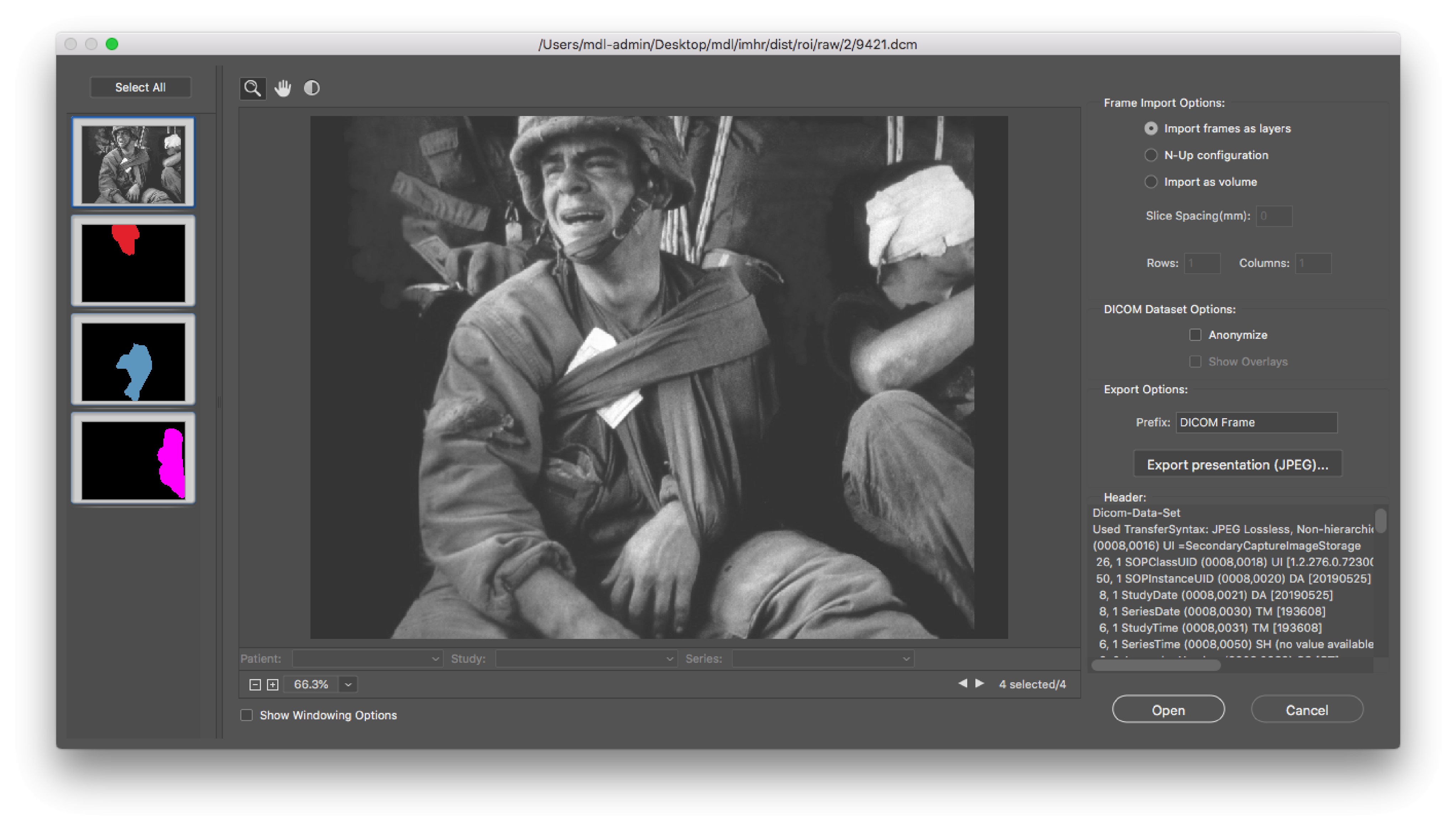
The recommended method of manually identifying each region of interest is by using a image media file that is able to store multiple layers of content. The two filetypes supported here are Adobe Photoshop Document (psd) and Digital Imaging and Communications in Medicine (dcm) filetypes. Each ROI can be stored as a seperate layer, which can then processed for exporting.
Figure: Example psd in Adobe Photoshop.
Figure: Example DICOM in Adobe Photoshop.
Haar Cascades¶
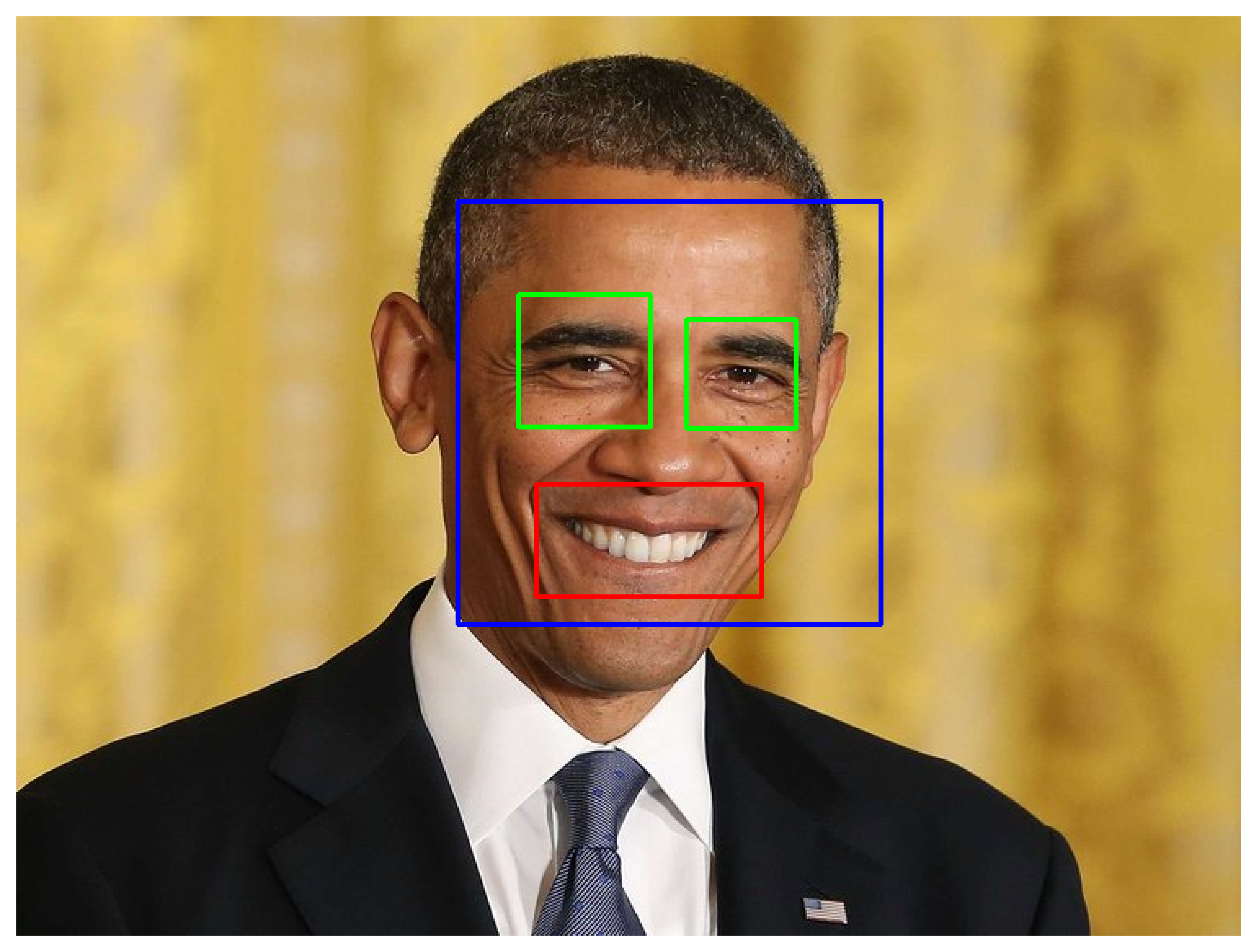
Another method for creating ROI’s is by using automated feature extraction using haar cascades feature classifiers. This is done by using machine-training to identify unique features (eyes, smile) from an image.
Figure: Example face, eye, and smile identfication using haar cascades.
Output¶
roi/
├── output/
│ ├── bounds.ias
│ ├── bounds.xlsx
│ ├── data/
│ │ ├── 2550_bounds.ias
│ │ └── 2550_bounds.xlsx
│ └── img/
│ ├── bounds/
│ │ ├── 2550.png
│ │ └── roi/
│ │ ├── 2550.1.png
│ │ └── 2550.2.png
│ ├── preprocessed/
│ │ └── 2550.png
│ └── raw/
│ └── 2550.png
└── raw/
├── 1/
│ ├── AM_201.psd
│ ├── AM_201.xcf
│ └── metadata.xlsx
└── 2/
├── 2550.psd
├── 2550.xcf
└── metadata.xlsx
After running the API, the following data will be outputted:
- Rerendered images (img)
- Dataviewer Region of Interest (ias): ‘./output/.ias’, ’./output/data/.ias’
- Data
- Dataviewer Region of Interest (ias): ‘./output/.ias’, ’./output/data/.ias’
- Excel Region of Interest (xlsx): ‘./output/.xlsx’, ’./output/data/.xlsx’
In addition, for each image, their corresponding ROIs will be drawn to a png file (./img/bounds). Individual ROIs are also drawn (‘./img/bounds/roi’).
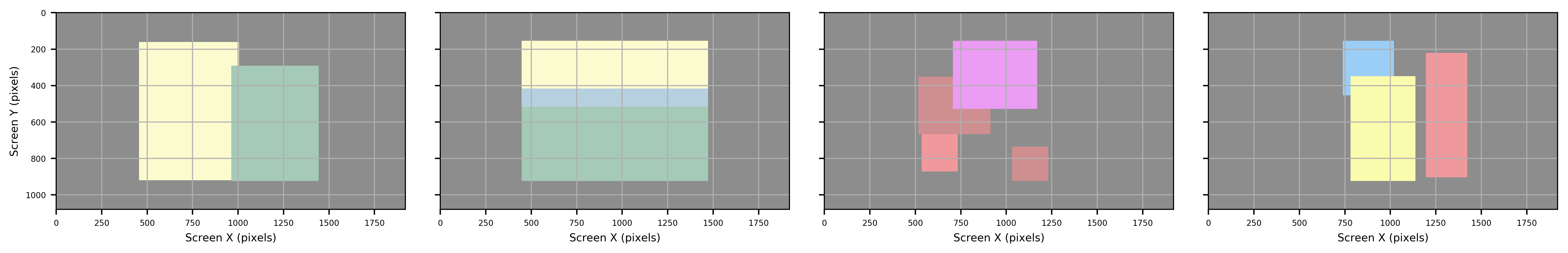
Bounds¶
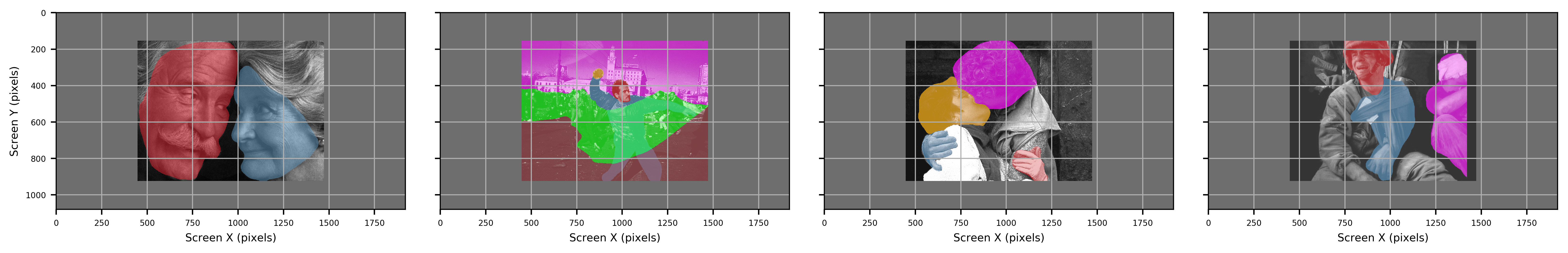
Bounds can be created for each region of interest with different levels of granularity. The list below provides an example of each, in order of granularity.
Figure: Original images before contour extraction.
Polygon
Figure: Contour Approximation (polygon) that will most closely match the orginal shape of the ROI.
Hull
Figure: Generated Convex Hull (hull), which is similar to but not as complex as a Contour Approximation and will include bulges for areas that are convex.
Circle
Figure: Circle creates a mininum enclosing circle, with the center equal to that of the ROI.
Rectangle (rotated)
Figure: Bounding Rectangle (rotated), with the position matching that of the general shape of the ROI.
Rectangle (straight)
Figure: Bounding Rectangle (straight) parallel to the shape of the background ima
Data¶
Two types of data are exported, contour shape and boundaries. Boundaries are saved in SR Research DataViewer (ias), and Microsoft Office Open XML Workbook (xlsx) formats, while contour shapes are saved in Hierarchical Data Format (HDF5; h5). Exported data can read either by SR Research DataViewer (ias) or any other data processing tool (xlsx, h5).
Note
Section 5.10.1 of Eyelink DataViewer Users Manual (3.2.1)
has more information about the ias format and how DataViewer can interpret it.
Both xlsx and HDF5 provide all metadata associated with each image, while ias files matches the format required by DataViewer. Of the three filetypes, HDF5 files are the most detailed providing both metadata for each ROI as well as exact pixel coordinates associated with each ROI.
For more information, click bounds.ias, bounds.xlsx, contours.h5 to see an example of each filetype.
Figure: Sample bounds output saved to xlsx.
[3]:
df
[3]:
| x0 | y0 | x1 | y1 | image | roi | id | shape_d | race | feature | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 937 | 554 | 792 | 422 | AM_201 | roi1 | 1 | RECTANGLE | asian | lefteye |
| 0 | 1118 | 552 | 973 | 420 | AM_201 | roi2 | 2 | RECTANGLE | asian | righteye |
| 0 | 1016 | 650 | 892 | 565 | AM_201 | roi3 | 3 | RECTANGLE | asian | nose |
| 0 | 880 | 756 | 1039 | 670 | AM_201 | roi4 | 4 | RECTANGLE | asian | mouth |
| 0 | 1114 | 862 | 818 | 762 | AM_201 | roi5 | 5 | RECTANGLE | asian | chin |
| 0 | 1109 | 407 | 801 | 307 | AM_201 | roi6 | 6 | RECTANGLE | asian | forehead |
| 0 | 783 | 665 | 704 | 532 | AM_201 | roi7 | 7 | RECTANGLE | asian | leftear |
| 0 | 1207 | 649 | 1144 | 494 | AM_201 | roi8 | 8 | RECTANGLE | asian | rightear |
| 0 | 938 | 524 | 785 | 409 | AM_203 | roi1 | 1 | RECTANGLE | asian | lefteye |
| 0 | 1125 | 525 | 976 | 409 | AM_203 | roi2 | 2 | RECTANGLE | asian | righteye |
| 0 | 1023 | 632 | 899 | 552 | AM_203 | roi3 | 3 | RECTANGLE | asian | nose |
| 0 | 1043 | 735 | 876 | 661 | AM_203 | roi4 | 4 | RECTANGLE | asian | mouth |
| 0 | 1040 | 837 | 862 | 759 | AM_203 | roi5 | 5 | RECTANGLE | asian | chin |
| 0 | 1137 | 382 | 778 | 259 | AM_203 | roi6 | 6 | RECTANGLE | asian | forehead |
| 0 | 766 | 637 | 693 | 474 | AM_203 | roi7 | 7 | RECTANGLE | asian | leftear |
| 0 | 1213 | 634 | 1147 | 462 | AM_203 | roi8 | 8 | RECTANGLE | asian | rightear |
| 0 | 928 | 538 | 778 | 425 | AM_204 | roi1 | 1 | RECTANGLE | asian | lefteye |
| 0 | 1124 | 535 | 975 | 427 | AM_204 | roi2 | 2 | RECTANGLE | asian | righteye |
| 0 | 1022 | 633 | 886 | 567 | AM_204 | roi3 | 3 | RECTANGLE | asian | nose |
| 0 | 862 | 735 | 1050 | 649 | AM_204 | roi4 | 4 | RECTANGLE | asian | mouth |
| 0 | 874 | 831 | 1038 | 755 | AM_204 | roi5 | 5 | RECTANGLE | asian | chin |
| 0 | 1137 | 410 | 776 | 262 | AM_204 | roi6 | 6 | RECTANGLE | asian | forehead |
| 0 | 766 | 658 | 700 | 483 | AM_204 | roi7 | 7 | RECTANGLE | asian | leftear |
| 0 | 1217 | 644 | 1148 | 483 | AM_204 | roi8 | 8 | RECTANGLE | asian | rightear |
| 0 | 944 | 531 | 792 | 406 | AM_223 | roi1 | 1 | RECTANGLE | asian | lefteye |
| 0 | 1132 | 524 | 983 | 405 | AM_223 | roi2 | 2 | RECTANGLE | asian | righteye |
| 0 | 1023 | 641 | 906 | 562 | AM_223 | roi3 | 3 | RECTANGLE | asian | nose |
| 0 | 1038 | 741 | 896 | 670 | AM_223 | roi4 | 4 | RECTANGLE | asian | mouth |
| 0 | 1053 | 847 | 875 | 776 | AM_223 | roi5 | 5 | RECTANGLE | asian | chin |
| 0 | 1150 | 386 | 778 | 275 | AM_223 | roi6 | 6 | RECTANGLE | asian | forehead |
| 0 | 775 | 641 | 695 | 488 | AM_223 | roi7 | 7 | RECTANGLE | asian | leftear |
| 0 | 1226 | 638 | 1145 | 483 | AM_223 | roi8 | 8 | RECTANGLE | asian | rightear |